Software
PDSFit
Description:
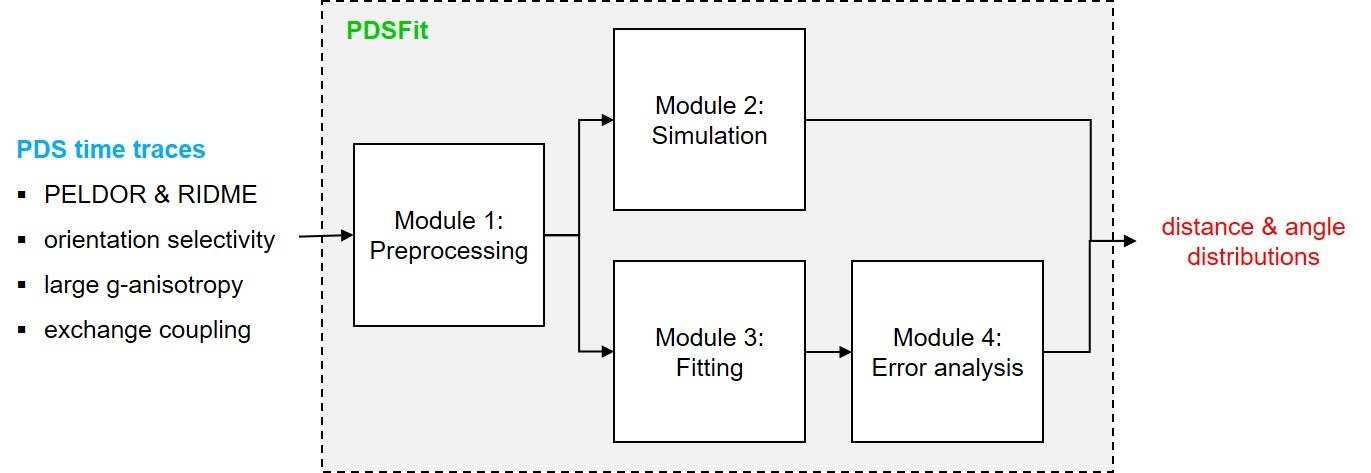
The program PDSFit allows the analysis of PELDOR and RIDME time traces with or without orientation selectivity. It can be applied to spin systems consisting of up to two spin centers with anisotropic g-factors and to spin systems with exchange coupling. It employs a model-based fitting of the time traces using parametrized distance and angular distributions, and parametrized PDS background functions.
Download:
• Source code
• Manual
Citation:
D. Abdullin, P. Rauh Corro, T. Hett, O. Schiemann, “PDSFit: PDS data analysis in
the presence of orientation selectivity, g-anisotropy, and exchange coupling”, Magn. Reson. Chem. 2024, 62, 37–60.
SnrCalculator
Description:
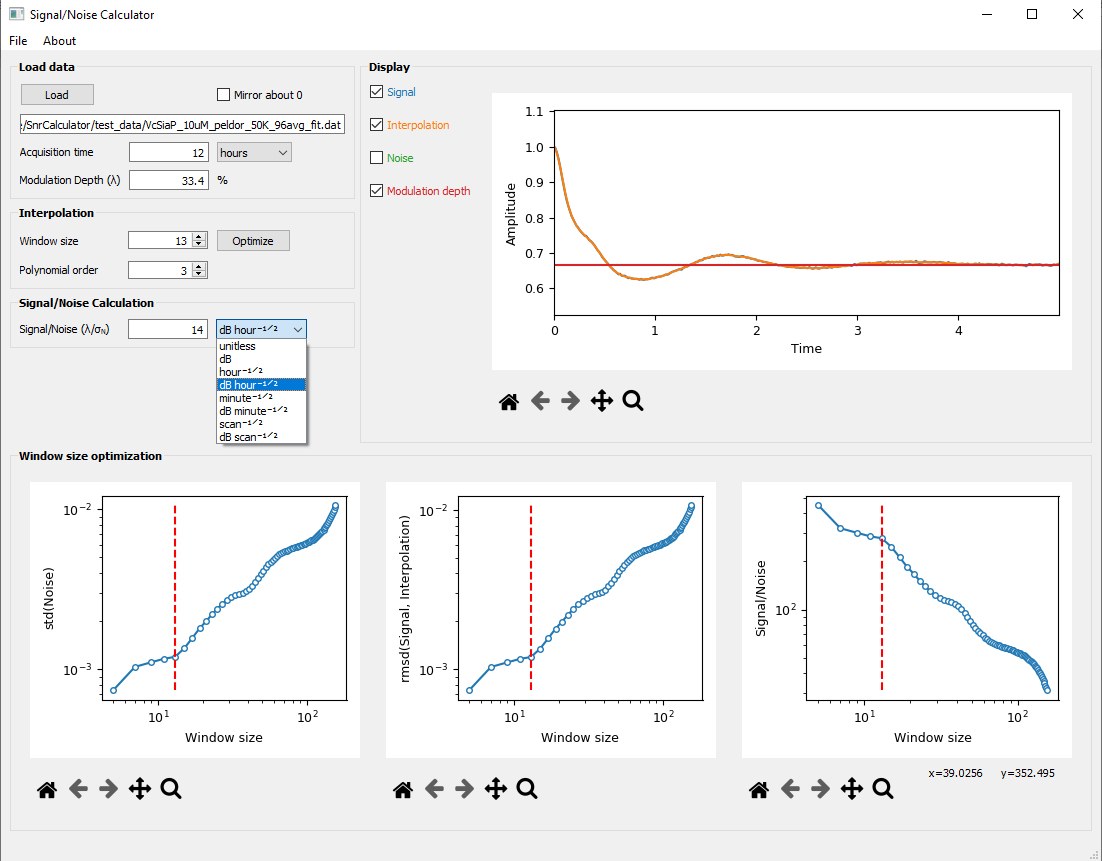
The program SnrCalculator allows determining the signal-to-noise ratio (SNR) of pulsed dipolar spectroscopy EPR (PDS) signals.
Download:
• Source code
• Manual (coming soon)
• Executables (Windows, Linux)
• GitHub
Citation:
D. Abdullin, P. Brehm, N. Fleck, S. Spicher, S. Grimme, O. Schiemann, “Pulsed EPR Dipolar Spectroscopy on Spin Pairs with one Highly Anisotropic Spin Center: The Low-Spin FeIII Case”, Chem. Eur. J. 2019, 25, 14388–14398 (see Supporting Information).
MtsslSuite
Description:
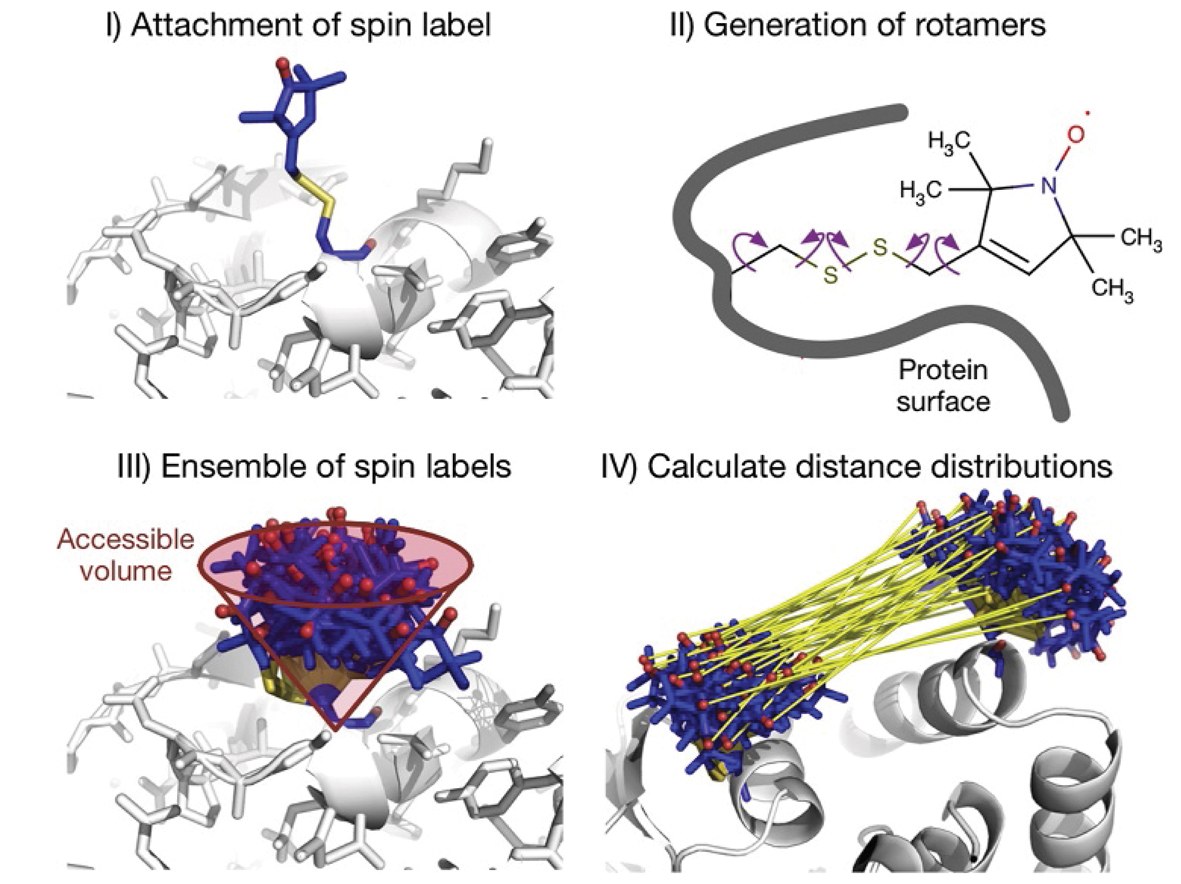
The software package contains four PyMOL plugins, MtsslWizard, MtsslTrilaterate, MtsslDock, and MtsslPlotter. MtsslWizard is a tool for in silico spin labeling of proteins and nucleic acids. MtsslTrilaterate allows performing the triangulation of intrinsic paramagnetic metal ions in macromolecules. MtsslDock allows performing the distance-constrained rigid-body docking.
Download:
https://pymolwiki.org/index.php/MtsslWizard (MtsslWizard)
https://pymolwiki.org/index.php/MtsslTrilaterate (MtsslTrilaterate)
https://pymolwiki.org/index.php/MtsslDock (MtsslDock)
Citation:
G. Hagelüken, R. Ward, J. H. Naimsith, O. Schiemann, “MtsslWizard: In silico Spin-Labelling and Generation of Distance Distributions in PyMOL", Appl. Magn. Reson. 2012, 42, 377-391.
G. Hagelueken, D. Abdullin, R. Ward, O. Schiemann, “In Silico Spin Labelling, Trilateration and Distance-Constrained Rigid Body Docking in PyMOL", Mol. Phys. 2013, 111, 2757-2766.
G. Hagelueken, D. Abdullin, O. Schiemann, “mtsslSuite: Probing biomolecular conformation by spin labeling studies", Methods Enzymol. 2015, 563, 595-622.
Note: The package is currently incompatible with the PyMOL versions >2.X. If you want to use MtsslSuite with the newest PyMOL, consider using the server version of MtsslSuite: http://www.mtsslsuite.isb.ukbonn.de